Welcome to NanoBind!
NanoBind is a comprehensive and efficient toolkit for nanobody-antigen binding prediction, offering five powerful models for a variety of interaction tasks.
We provide a user-friendly and robust webserver, enabling researchers to easily predict binding sites, affinities, and interaction strengths using FASTA sequences. The simple usage tutorial is as follows:
-
NanoBind_Example data display:

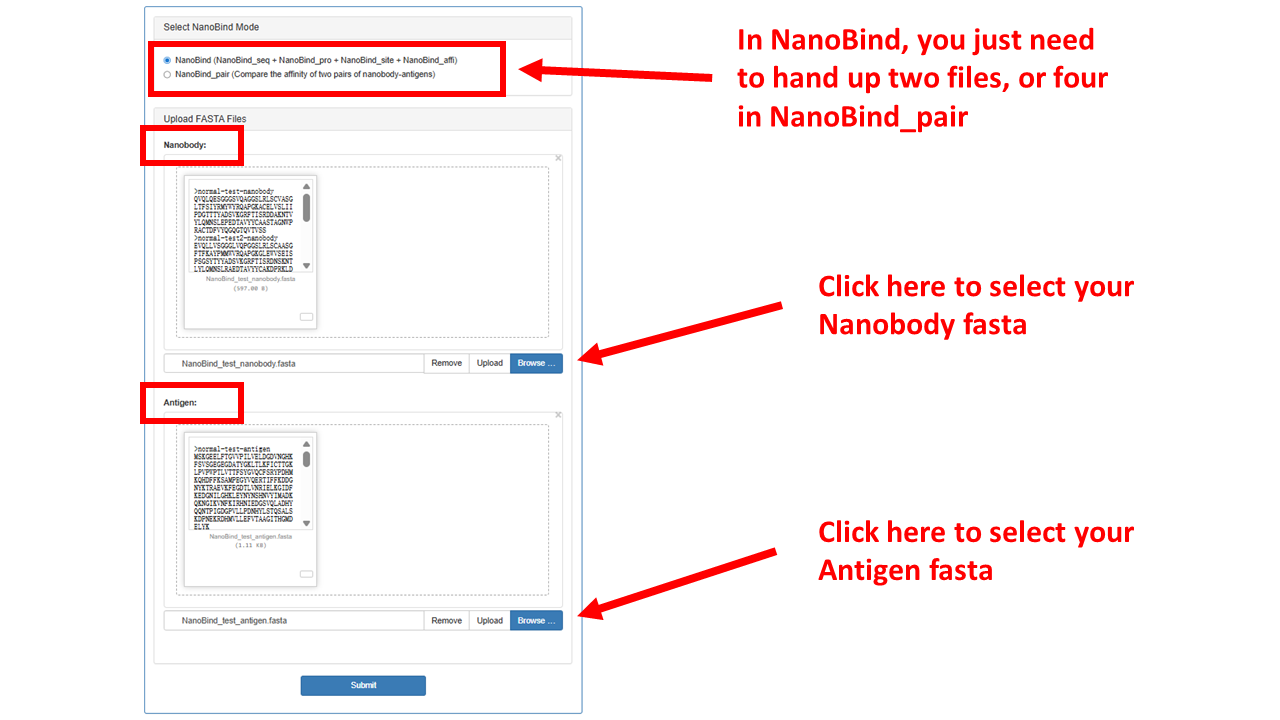
- Choose the prediction mode:

- Upload the required FASTA files:
All uploaded files must be in FASTA format. Please upload the nanobody FASTA file in the Nanobody section
and the antigen FASTA file in the Antigen section. NanoBind will read the sequences from both files in order
and match them one-to-one for prediction.
Note: NanoBind will automatically check for invalid input formats. However, it cannot detect mismatches in the
sequence order between the two files. Therefore, it is critical to ensure that the sequences in both files are
listed in the same order, with each nanobody properly aligned to its corresponding antigen.
- Submit:
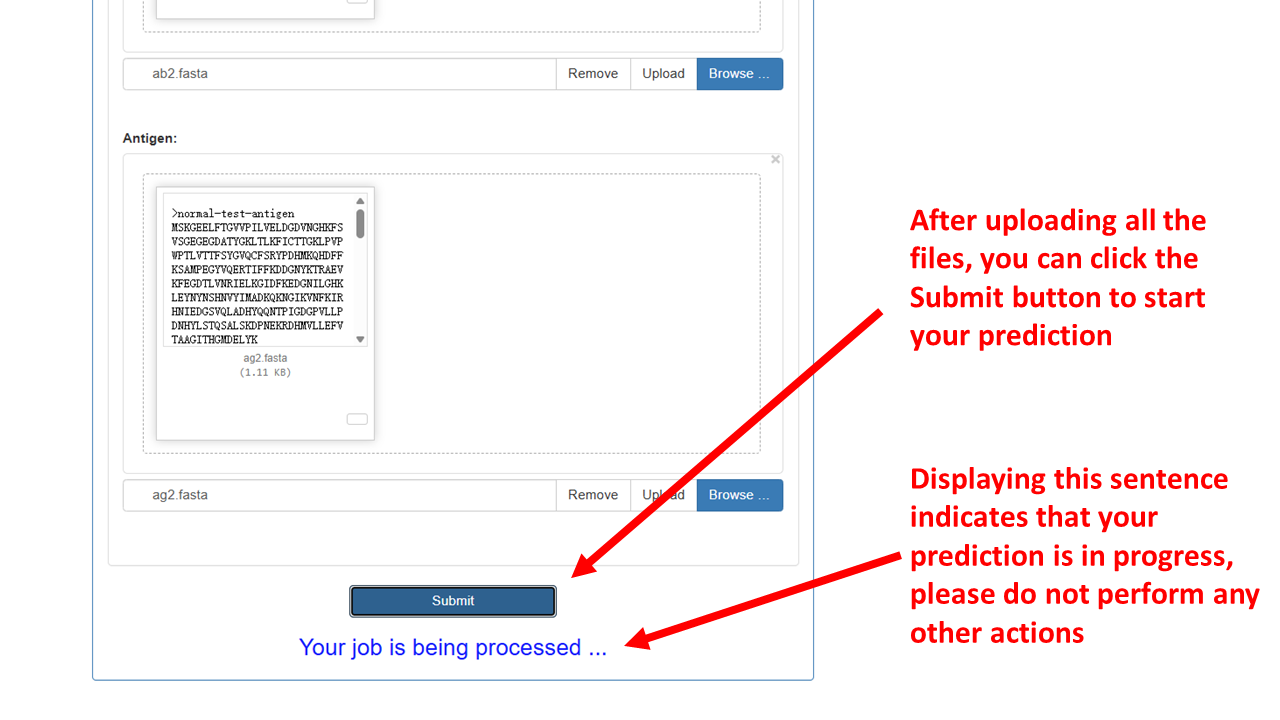
-
Interpretation of Results:
In NanoBind mode, you will receive the results shown in the following figure.

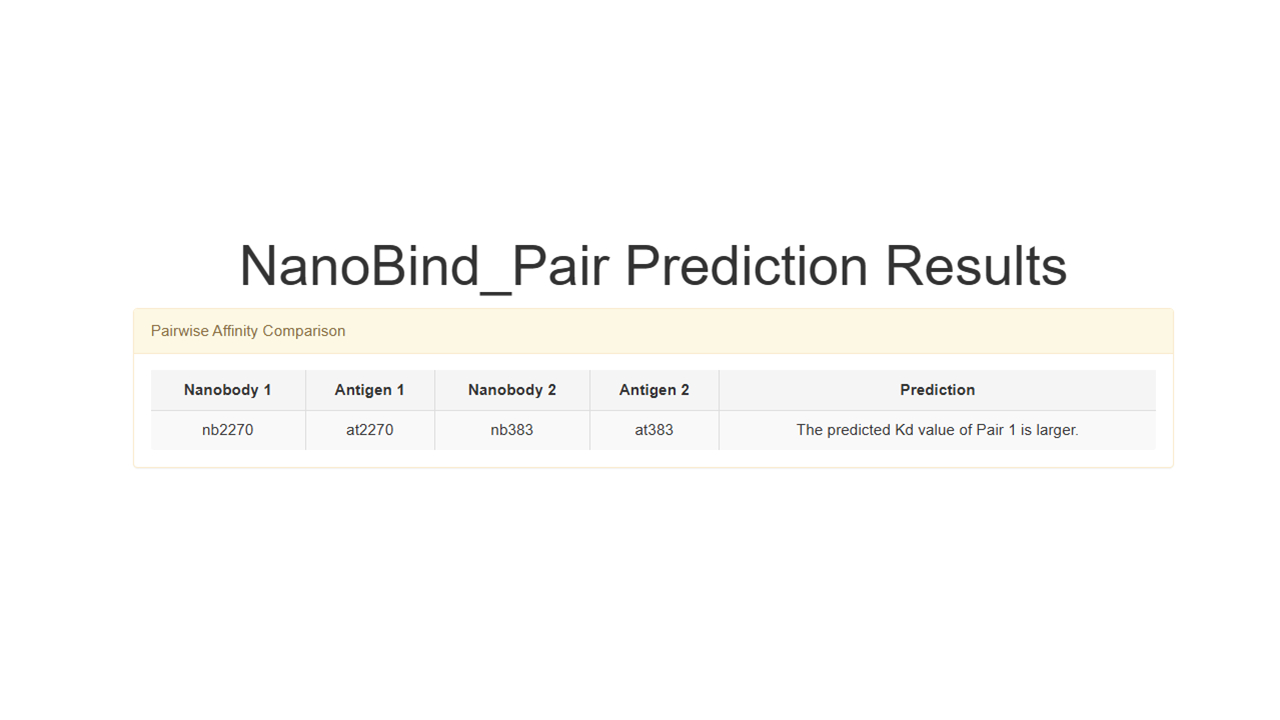
In NanoBind_pair mode, you will receive the results shown in the following figure.
- NanoBind_Example data display:
- Choose the prediction mode:
- Upload the required FASTA files:
All uploaded files must be in FASTA format. Please upload the nanobody FASTA file in the Nanobody section and the antigen FASTA file in the Antigen section. NanoBind will read the sequences from both files in order and match them one-to-one for prediction.
Note: NanoBind will automatically check for invalid input formats. However, it cannot detect mismatches in the sequence order between the two files. Therefore, it is critical to ensure that the sequences in both files are listed in the same order, with each nanobody properly aligned to its corresponding antigen. - Submit:
-
Interpretation of Results:
In NanoBind mode, you will receive the results shown in the following figure.

In NanoBind_pair mode, you will receive the results shown in the following figure.





If you think NanoBind is useful, please kindly cite the following paper:
Webserver update:
This work is
openly licensed via CC0 1.0