Welcome to PepNet!
PepNet is a sequence-based deep learning model for the prediction of both AIPs and AMPs
We provides a convenient and efficient PepNet webserver for researchers to predict the antimicrobial or anti-inflammatory activity of peptide sequences.
Fast mode:
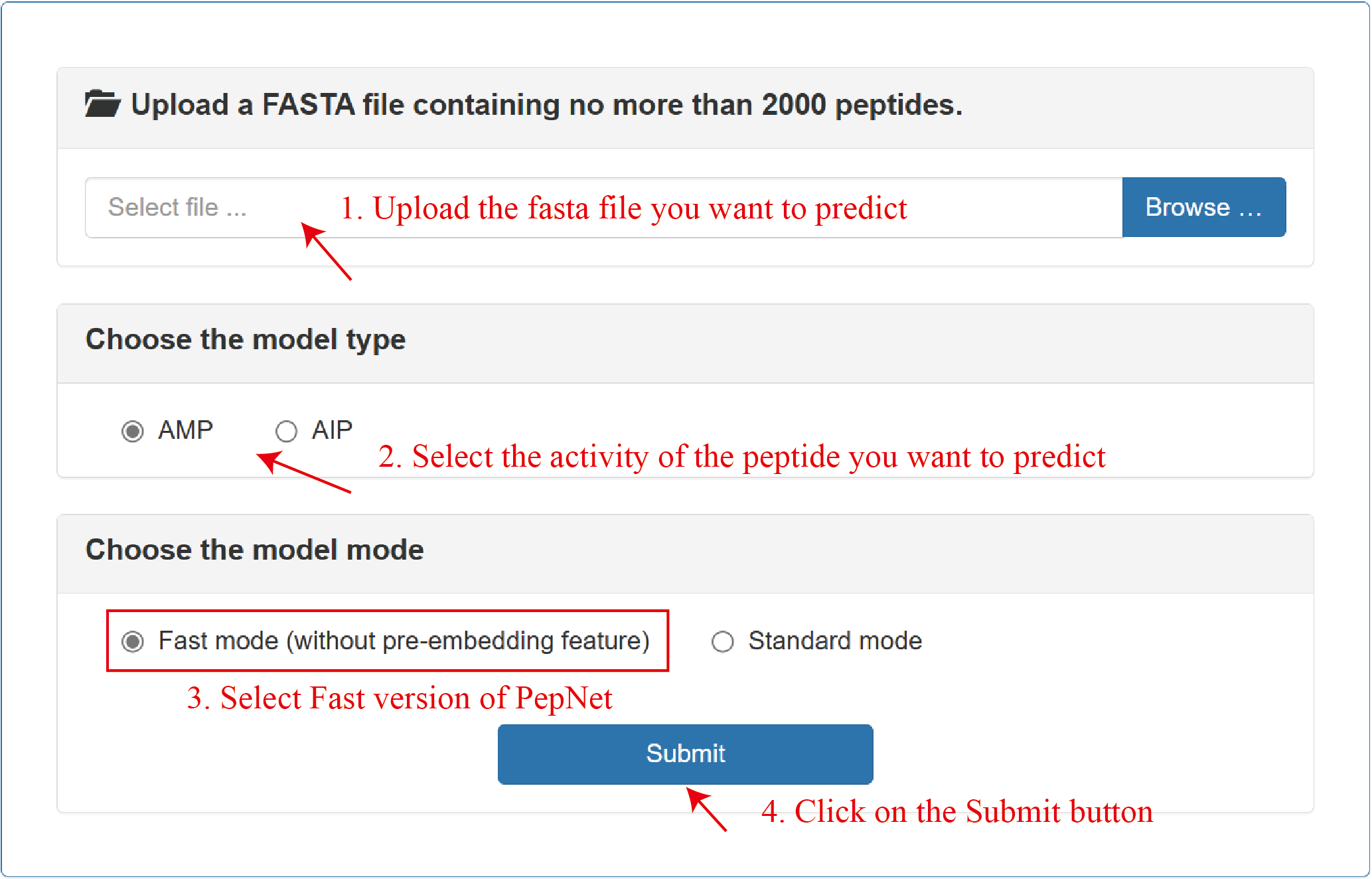
Standard mode:
- Create an environment
- # create $ conda create -n PepNet python=3.8
- # activate $ source activate PepNet
- Install PepNet dependencies
- (1) Install pytorch 2.4.0 (For more details, please refer to https://pytorch.org/) For linux:
- (2) Install the requirements. For linux:
- (3) Extraction the pretrained features generated by ProtT5-XL-U50
- (1) Download the source code of ProtTrans from https://github.com/agemagician/ProtTrans.
- (2) Download the ProtT5-XL-U50 model from https://huggingface.co/Rostlab/prot_t5_xl_uniref50/tree/main.
- (3) Place the downloaded model files in this directory {ProtTrans_path}/Embedding/Rostlab/prot_t5_xl_half_uniref50-enc
- (4) Generate the pretrained feature for a fasta file. $ cd {ProtTrans_path}/Embedding
- Standard mode using
# CUDA 11.8
$ pip install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
$ pip install h5py==3.11.0
$ pip install transformers==4.43.2
$ python prott5_embedder.py --input {the path of the fasta file} --output {the path of the output H5PY file, e.g, out.h5} --model {the path of the ProtT5-XL-U50 model you download}

If you think PepNet is useful, please kindly cite the following paper:
Webserver update:
May, 13th, 2024: the first version of PepNet server was established.
This work is
openly licensed via CC0 1.0