Welcome to PepNet!
Identifying anti-inflammatory peptides (AIPs) and antimicrobial peptides (AMPs) is crucial for the discovery of innovative and effective peptide-based therapies targeting inflammation and microbial infections. PepNet is a sequence-based deep learning model for the prediction of both AIPs and AMPs
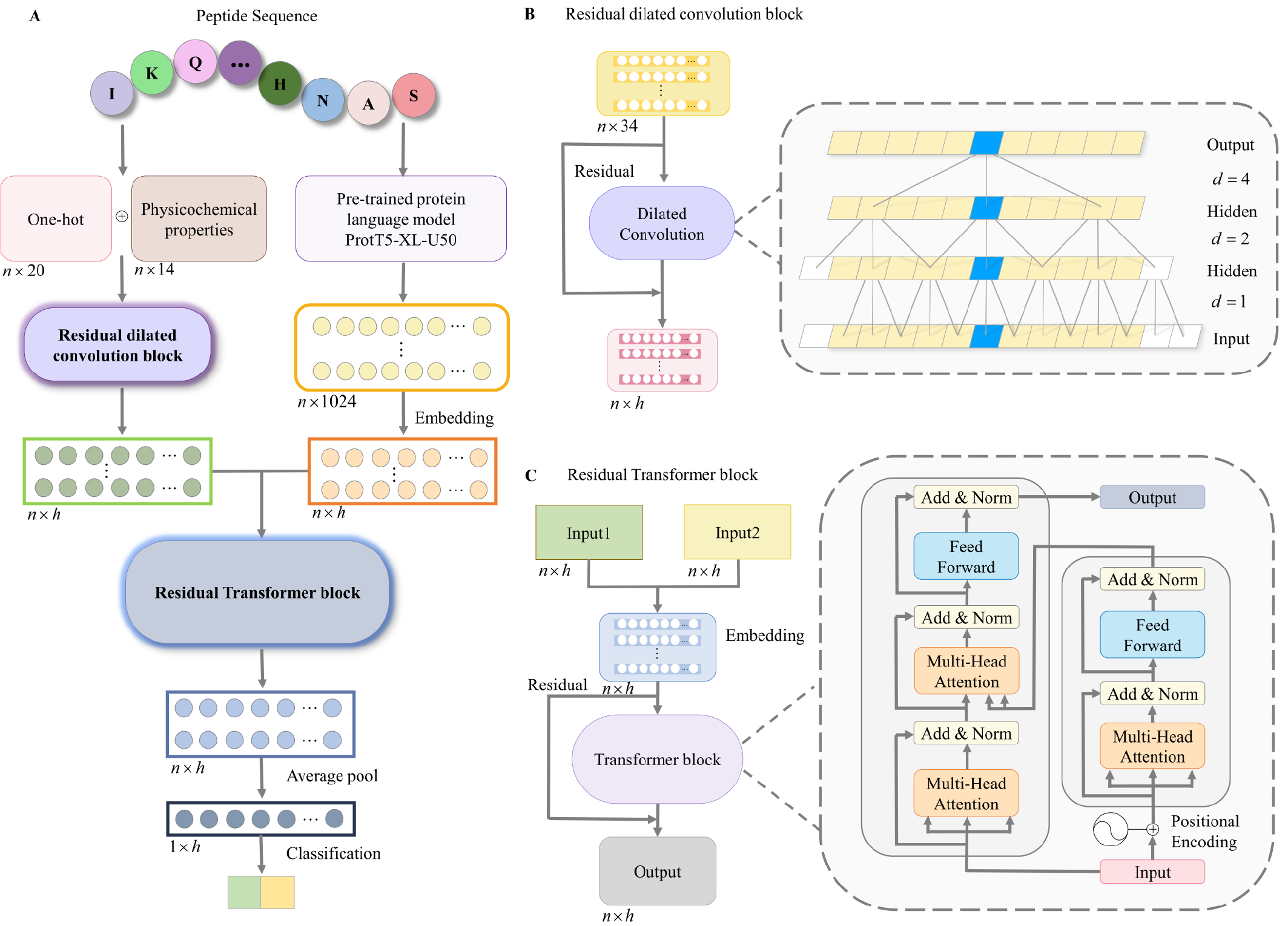
The framework first extracts the type and physicochemical properties of amino acids, which are fed into a residual dilated convolution block to capture the information from variable spaced sequence neighbors progressively. Furthermore, along with the reused pre-embedding features from a protein large language model, the sequence feature map is transformed via a residual Transformer block to capture the information from all positional residues. Finally, the feature map is average pool for generation a sequence representation to classify the peptide.
We provides a convenient and efficient PepNet webserver for researchers to predict the antimicrobial or anti-inflammatory activity of peptide sequences. You can also use the source code of PepNet which is provided on our Github page http:.
If you think PepNet is useful, please kindly cite the following paper:
Webserver update:
May, 13th, 2024: the first version of PepNet server was established.
This work is
openly licensed via CC0 1.0