Welcome to GraphRBF!
Predicting protein-protein and protein-nucleic acid interaction sites from protein structures can provide insights into biological processes related to the function of proteins and provide key technical guidance for disease diagnosis and the design of new drugs.
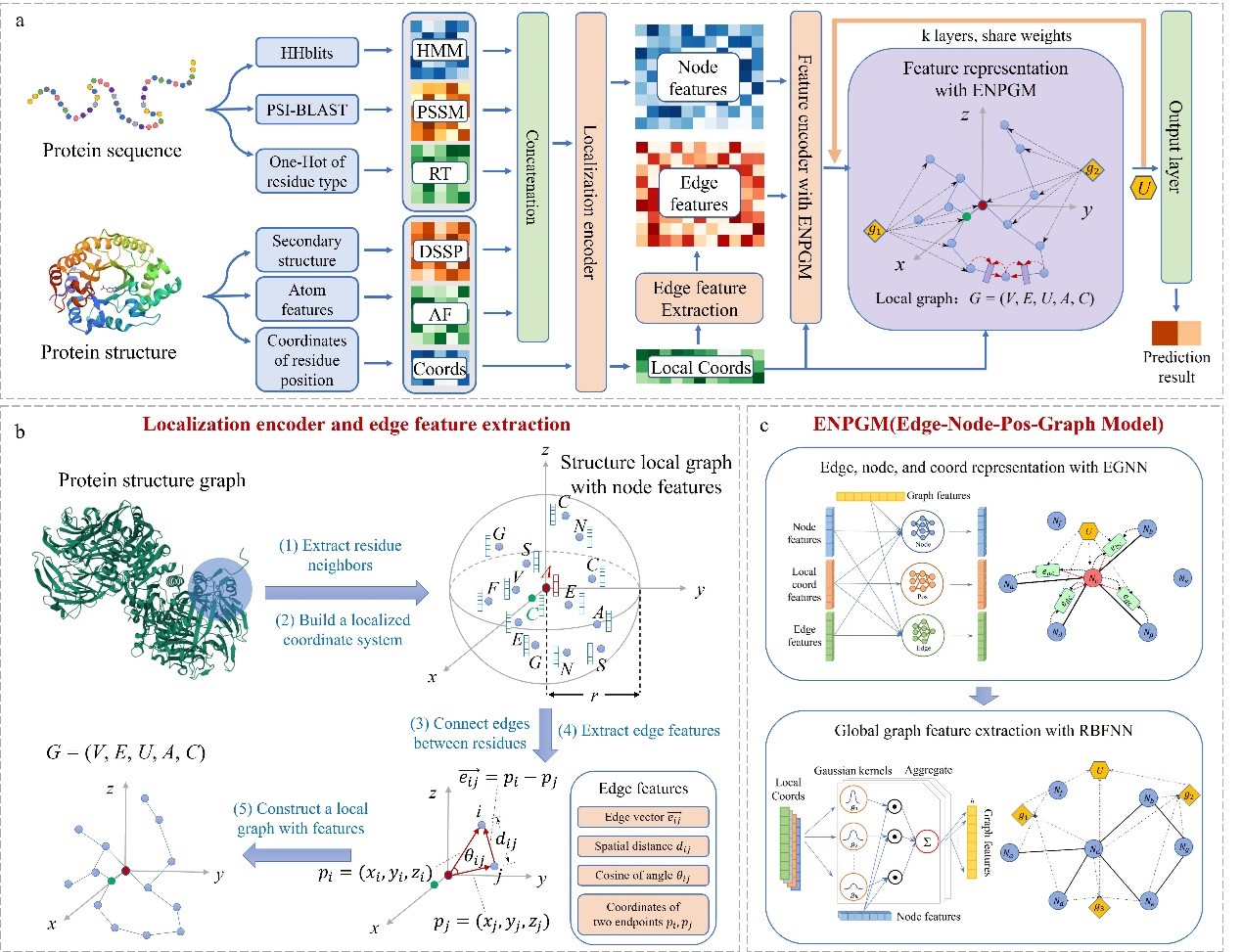
In the present invention, we designed GraphRBF, an end-to-end interpretable hierarchical geometric deep learning model. GraphRBF constructs local neighbor representations of target residues based on the spatial-chemical arrangement of amino acid neighbors, which ensures translational and rotational invariance. After that, we combine the equivariant graph neural network and radial basis function neural network to extract features directly from the local 3D structure of proteins, embedding the hidden layer feature representation of amino acids.
Users can use their own data to predict the driver gene for a particular cancer through our online GraphRBF webserver. Source code of our server is provided on our Github page https://github.com/Wssduer/GraphRBF.
If you think GraphRBF is useful, please kindly cite the following paper:
Webserver update:
May, 10th, 2023: the first version of GraphRBF server was established.
This work is
openly licensed via CC0 1.0