Welcome to GeoNet!
GeoNet is a powerful and interpretable geometric deep learning model for identifying protein interaction sites based on protein structure.
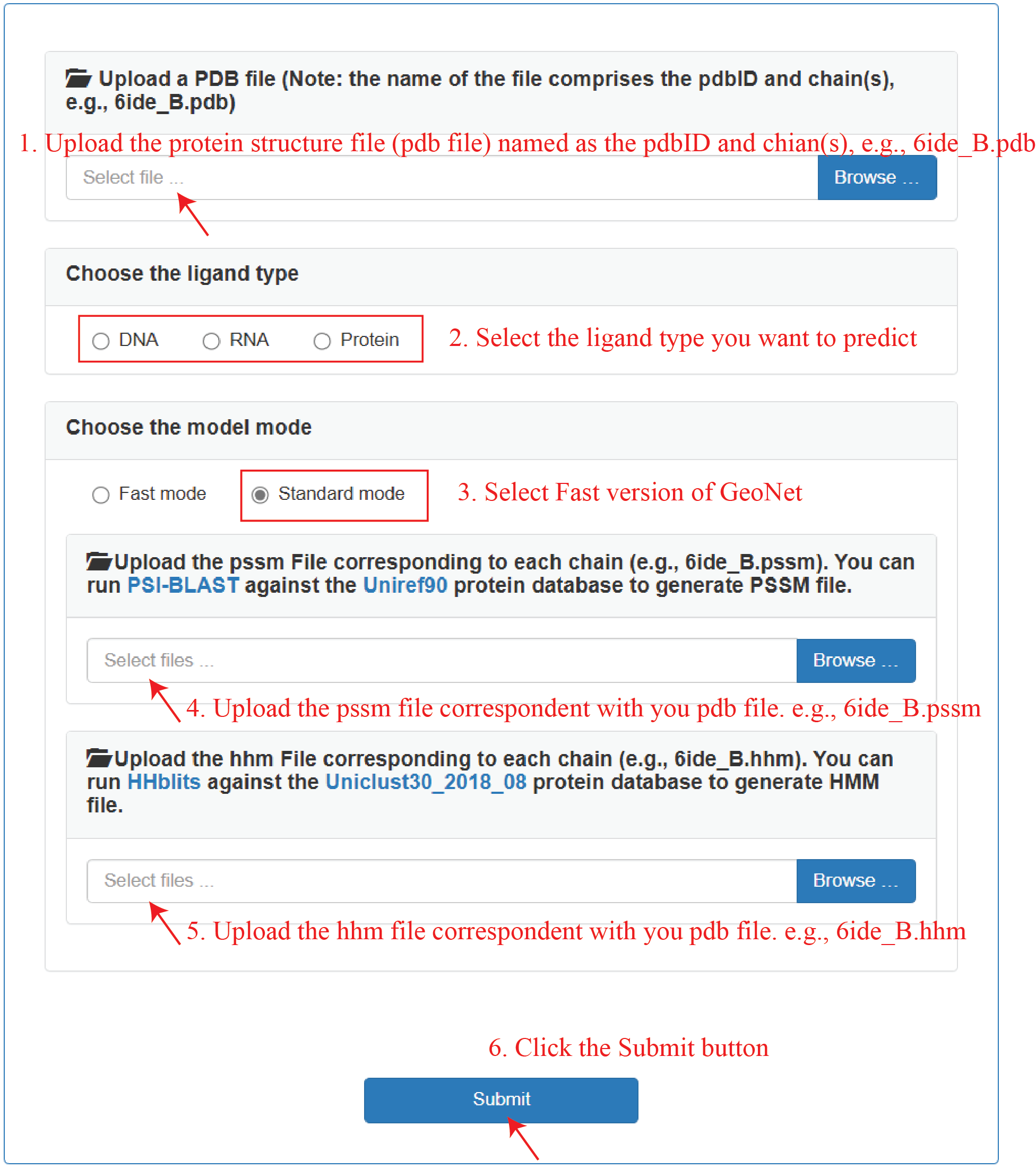
We provides a convenient and efficient GeoNet webserver for researchers to predict the protein-DNA, protein-RNA, and protein-protein binding sites.
Fast mode:

Standard mode:
- Install blast+ for extracting PSSM (position-specific scoring matrix) profiles
- To install ncbi-blast-2.14.0+ or latest version and download NR database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) for psiblast, please refer to BLAST® Help (https://www.ncbi.nlm.nih.gov/books/NBK52640/). $ tar zxvpf ncbi-blast-2.14.0.tar.gz
- Set the absolute paths of blast+ and NR databases in the script "scripts/prediction.py". $ PSIBLAST = '/mnt/data0/Hanjy/software/ncbi-blast-2.14.0+/bin/psiblast'
- Install HHblits for extracting HMM profiles
- To install HHblits and download uniclust30_2018_08 (http://wwwuser.gwdg.de/~compbiol/uniclust/2018_08/uniclust30_2018_08_hhsuite.tar.gz). For HHblits, please refer to https://github.com/soedinglab/hh-suite. $ git clone https://github.com/soedinglab/hh-suite.git
- Set the absolute paths of HHblits and uniclust30_2018_08 databases in the script "scripts/prediction.py". $ HHblits = '/mnt/data0/Hanjy/software/hh-suite/build/bin/hhblits'
- Install DSSP for extracting SS (Secondary structure) profiles
- DSSP is contained in "scripts/dssp", and it should be given executable permission by: $ chmod +x scripts/dssp
- Standard mode using
$ PSIBLAST_DB = '/mnt/data0/Hanjy/software/database/uniref90/uniref90'
$ mkdir -p hh-suite/build && cd hh-suite/build
$ cmake -DCMAKE_INSTALL_PREFIX=. ..
$ make -j 4 && make install
$ export PATH="$(pwd)/bin:$(pwd)/scripts:$PATH"
$ HHblits_DB = '/mnt/data0/Hanjy/software/database/uniclust30_2018_08/uniclust30_2018_08'
If you think GeoNet is useful, please kindly cite the following paper:
Webserver update:
May, 13th, 2024: the first version of GeoNet server was established.
This work is
openly licensed via CC0 1.0